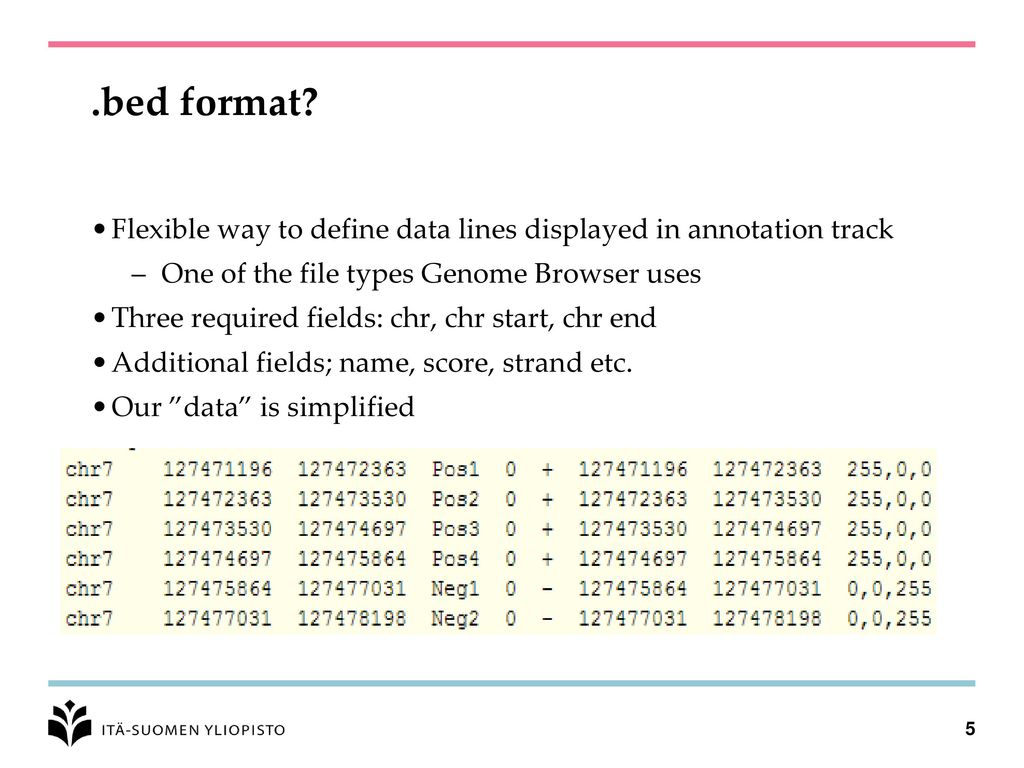
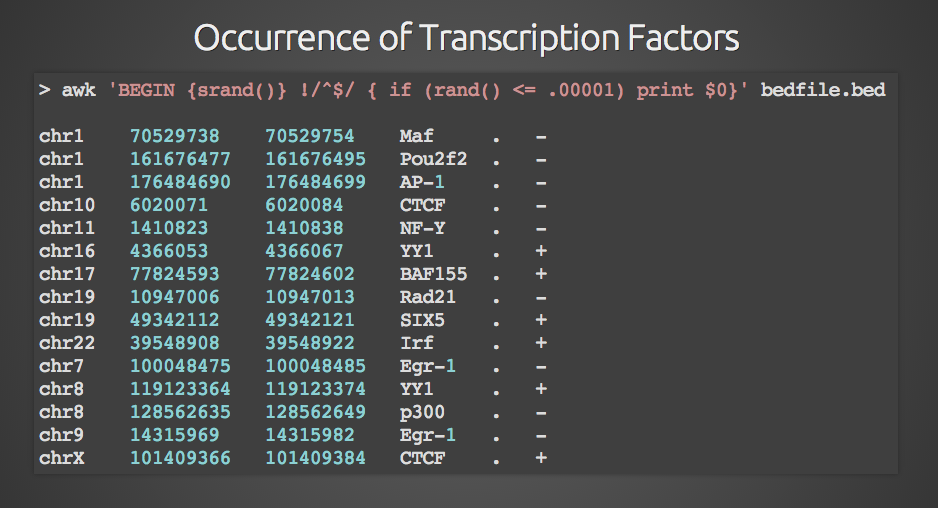
Example RNAcentral BED file (note the last two columns containing RNA... | Download Scientific Diagram

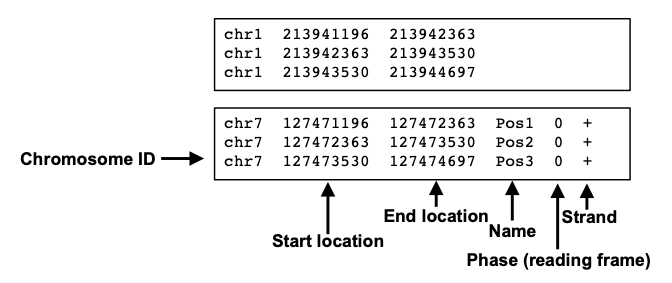
BED file examples. RefSeq transcript annotation in BED format on genome... | Download Scientific Diagram

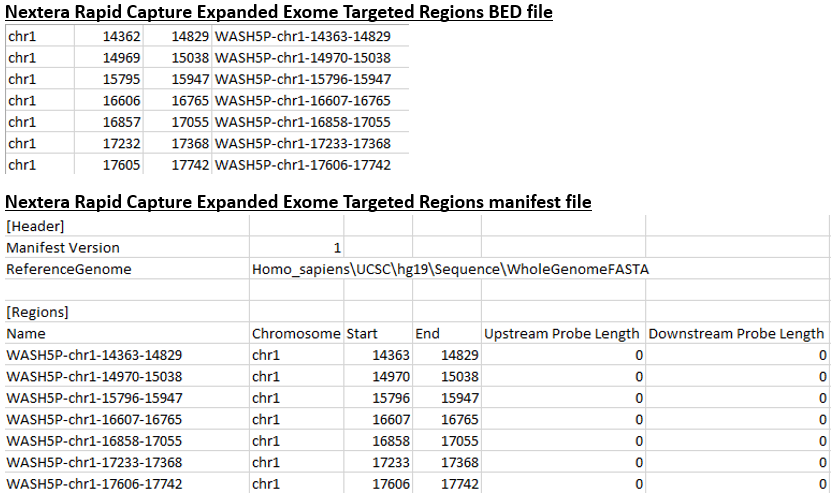
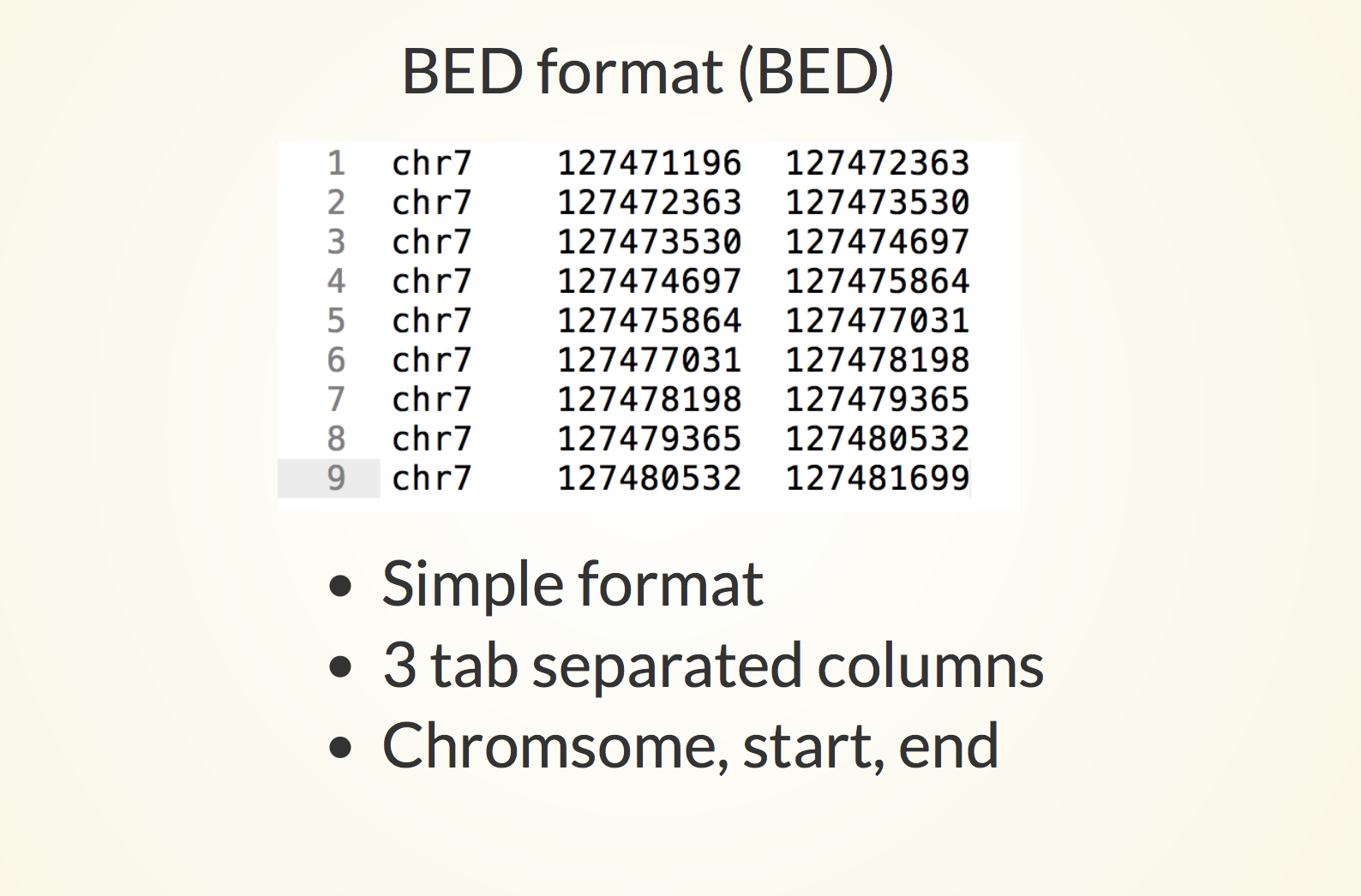
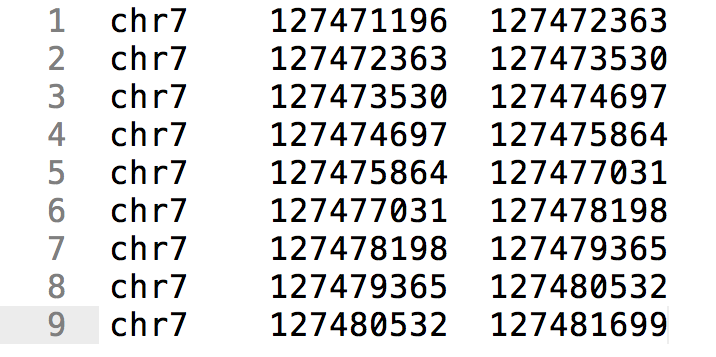
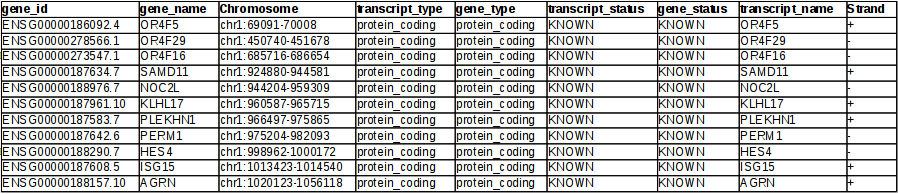
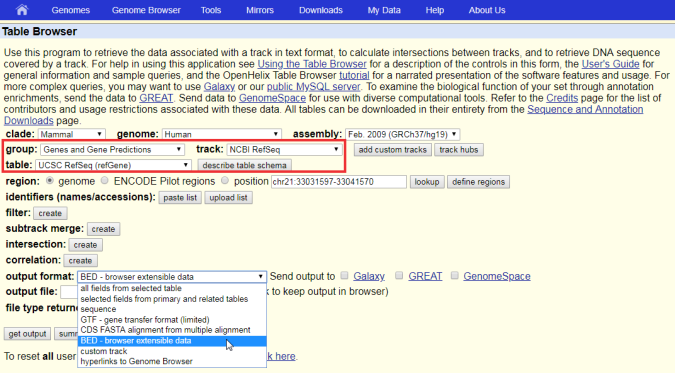
Genome Browser - Help | Gene Expression Profiling | eArray | Custom Microarrays | Gene Expression | ArrayGen